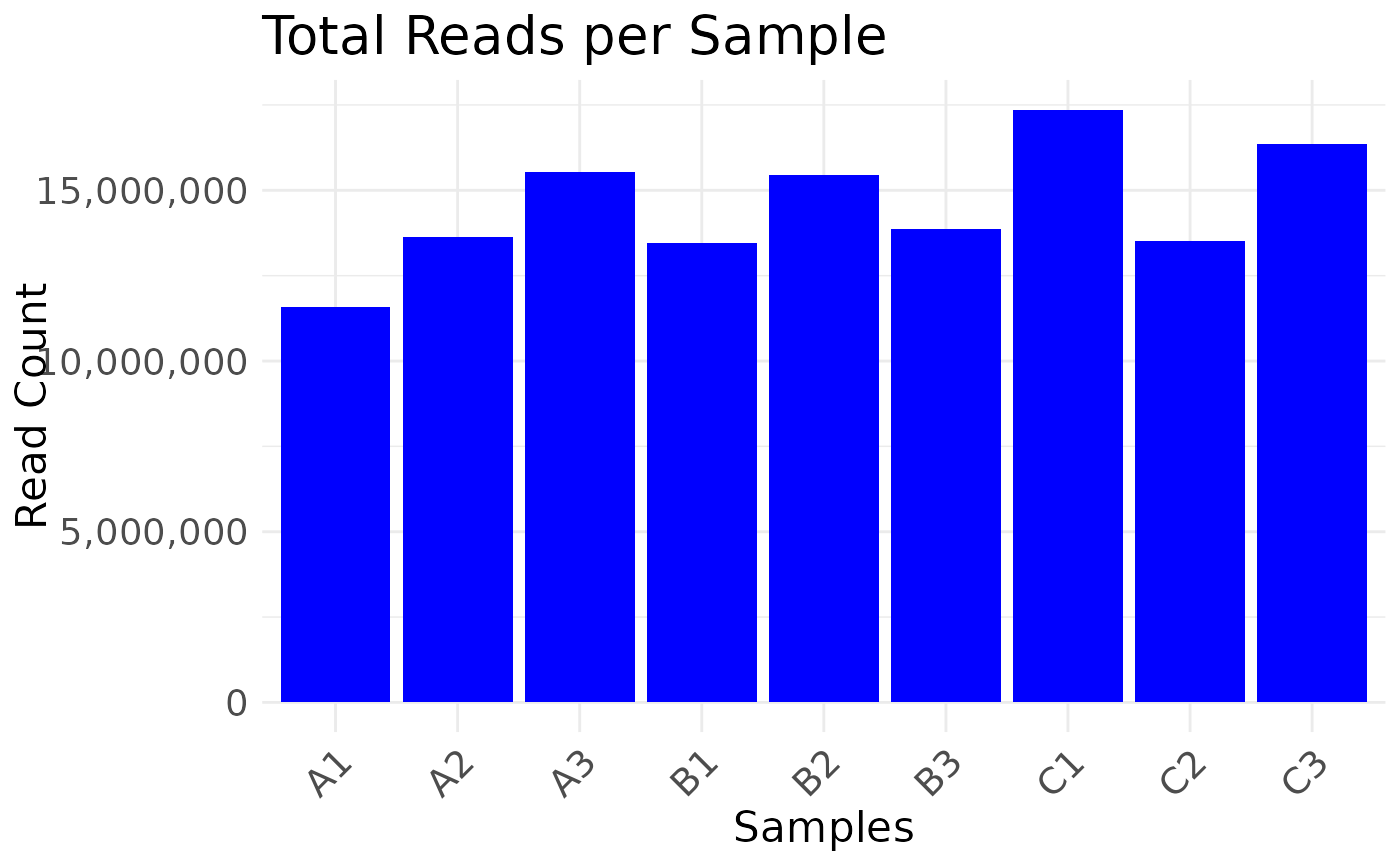
Plot read depth for multiOmicDataSet
Arguments
- moo_counts
multiOmicDataSetcontainingcount_type&sub_count_typein the counts slot- count_type
the type of counts to use. Must be a name in the counts slot (
names(moo@counts)).- sub_count_type
used if
count_typeis a list in the counts slot: specify the sub count type within the list. Must be a name innames(moo@counts[[count_type]]).
See also
plot_read_depth generic
Other plotters for multiOmicDataSets:
plot_corr_heatmap_moo,
plot_histogram_moo,
plot_pca_moo
Examples
# multiOmicDataSet
moo <- multiOmicDataSet(
sample_metadata = nidap_sample_metadata,
anno_dat = data.frame(),
counts_lst = list(
"raw" = nidap_raw_counts,
"clean" = nidap_clean_raw_counts
)
)
plot_read_depth(moo, count_type = "clean")